These research programs are in close collaboration with our clinical partners in particular Prof. Carsten Müller-Tidow (Heidelberg University Hospital) and Prof. Wolf-Karsten Hofmann (University Medical Centre Mannheim). Moreover, we have recently founded the Heidelberg Leukemia Network (HeLeNe) to integrate all Heidelberg groups working on basic, translational and clinical aspects of leukemia. The goal is to push forward innovative translational developments and implement them in a clinical setting. Together with reverse translational approaches we are establishing a powerful integrated network to improve outcome of AML patients by increased mechanistic insights into the disease.
Myelodysplastic Syndromes (MDS)
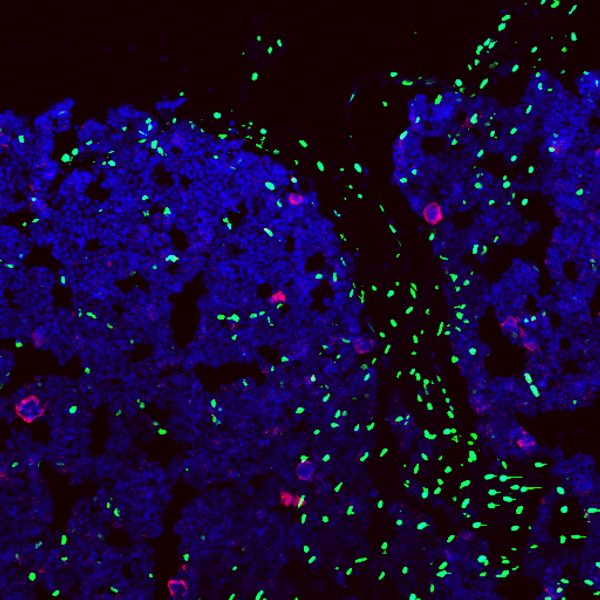
We have recently reported, that malignant progenitors isolated from MDS patients reprogram their direct mesenchymal microenvironment in the bone marrow to form a “MDS-stem cell niche unit”, which after transplantation as a whole can re-initiate the disease in immuno-deficient recipient mice (Medyouf H. et al., Cell Stem Cell 2014).
Metabolism meets the epigenome in AML stem cells
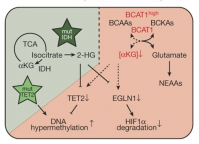 We run a program analyzing leukemic stem cells (LSC) by multi-omics analysis (methylome, transcriptome and proteome, see Raffel et al., Nature, 2017). This analysis revealed that branched chain amino acid (BCAA) catalysis and its key enzyme BCAA transaminase 1 (BCAT1) is specifically overexpressed in LSCs versus leukemia blast cells. We then showed that BCAT1 is essential for the functional activity of AML stem cells. BCAT1 links BCAA catabolism to the control of intracellular α-ketoglutarate (αKG) level. We found that the BCAA-BCAT-αKG pathway mimics the effects of IDH and TET2 mutations and is associated with poor overall survival in IDHwtTET2wt AML patients. Our data suggest that suppression of αKG-dependent dioxygenases constitutes a common feature of AML and, next to mutations in IDH and TET2, the identified BCAA-BCAT1-αKG axis is an alternative, non-genetic mechanism in LSCs to control the activity of these enzymes (Raffel et al., Nature 2017). Our data not only identify a novel signaling node, but also linking LSC metabolism to epigenetic control of the LSC epigenome (Figure 4) (Raffel et al., Nature, 2017).
We run a program analyzing leukemic stem cells (LSC) by multi-omics analysis (methylome, transcriptome and proteome, see Raffel et al., Nature, 2017). This analysis revealed that branched chain amino acid (BCAA) catalysis and its key enzyme BCAA transaminase 1 (BCAT1) is specifically overexpressed in LSCs versus leukemia blast cells. We then showed that BCAT1 is essential for the functional activity of AML stem cells. BCAT1 links BCAA catabolism to the control of intracellular α-ketoglutarate (αKG) level. We found that the BCAA-BCAT-αKG pathway mimics the effects of IDH and TET2 mutations and is associated with poor overall survival in IDHwtTET2wt AML patients. Our data suggest that suppression of αKG-dependent dioxygenases constitutes a common feature of AML and, next to mutations in IDH and TET2, the identified BCAA-BCAT1-αKG axis is an alternative, non-genetic mechanism in LSCs to control the activity of these enzymes (Raffel et al., Nature 2017). Our data not only identify a novel signaling node, but also linking LSC metabolism to epigenetic control of the LSC epigenome (Figure 4) (Raffel et al., Nature, 2017).
Super-Enhancers in AML
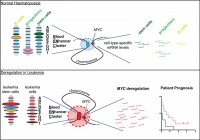
As described, above, we showed that Myc is regulated by a large modular gene enhancer cluster (super-enhancer) during normal hematopoiesis and in leukemia. Most strikingly, we could show in collaboration with the laboratory of Dr. John Dick (Toronto), that the activity of some sub-enhancers normally used in HSCs are altered in AML stem cells and this correlates with response to chemotherapy and patient survival (Bahr et al., Nature, 2018).
Minimal Residual Disease in AML
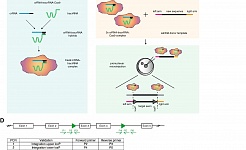
We have recently started a major program to study minimal residual disease (MRD) in leukemia patients. Resistant cells are retained after the initially successful chemotherapy regime and if such resistant cells have leukemic stem cell activity, they will be able to restart the leukemia leading to relapse and eventually death. We will combine state of the art technologies including next generation sequencing with single-cell approaches to identify such rare cells in patients, identify the mechanism of therapy resistance and develop novel strategies to efficiently eliminate such cells to prevent relapse in AML. For this program we are closely interacting with the Team of Dr. Carsten Müller-Tidow (Heidelberg University Hospital) and other University hospitals in Germany to develop novel tools to diagnose and target therapy resistance (stem) cells with the goal to improve the outcome of AML patients.
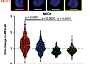
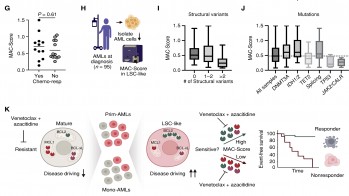 We identified leukemic stem cells (LSC) as primary targets of 5-AZA/VEN whose elimination determined the therapy outcome. LSCs of 5-AZA/VEN-refractory patients displayed perturbed apoptotic dependencies. We developed and validated a flow cytometry-based "Mediators of apoptosis combinatorial score" (MAC-Score) linking the ratio of protein expression of BCL2, BCL-xL, and MCL1 in LSCs. MAC scoring predicts initial response with a positive predictive value of more than 97% associated with increased event-free survival. In summary, combinatorial levels of BCL2 family members in AML-LSCs are a key denominator of response, and MAC scoring reliably predicts patient response to 5-AZA/VEN (Waclawiczek et al., Cancer Discovery, 2023).
We identified leukemic stem cells (LSC) as primary targets of 5-AZA/VEN whose elimination determined the therapy outcome. LSCs of 5-AZA/VEN-refractory patients displayed perturbed apoptotic dependencies. We developed and validated a flow cytometry-based "Mediators of apoptosis combinatorial score" (MAC-Score) linking the ratio of protein expression of BCL2, BCL-xL, and MCL1 in LSCs. MAC scoring predicts initial response with a positive predictive value of more than 97% associated with increased event-free survival. In summary, combinatorial levels of BCL2 family members in AML-LSCs are a key denominator of response, and MAC scoring reliably predicts patient response to 5-AZA/VEN (Waclawiczek et al., Cancer Discovery, 2023).